Figure 2 from Mass Spectrometry-Based Identification of Muscle-Associated and Muscle-Derived Proteomic Biomarkers of Dystrophinopathies. | Semantic Scholar

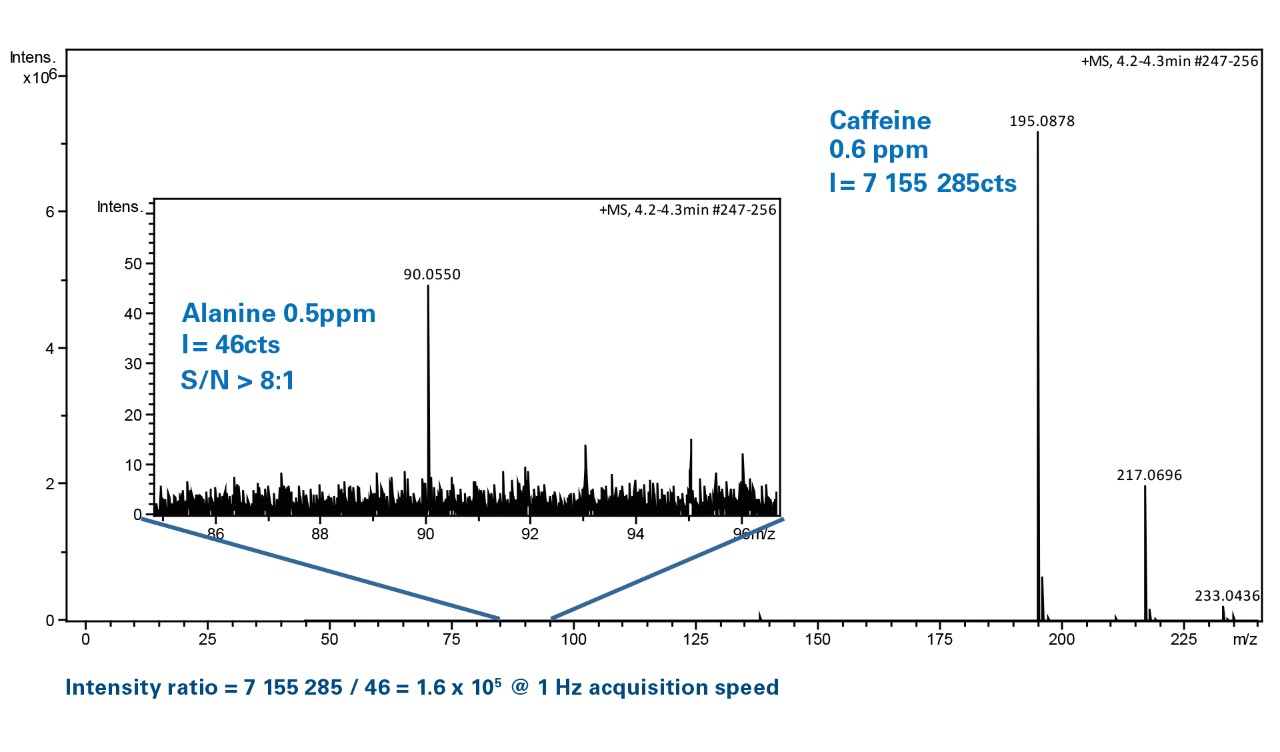
Expanding the linear dynamic range for quantitative liquid chromatography-high resolution mass spectrometry utilizing natural isotopologue signals - ScienceDirect
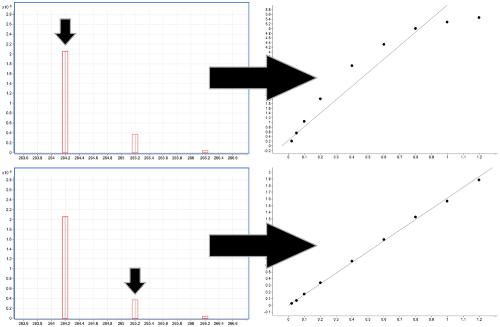
Increasing the linear dynamic range in LC–MS: is it valid to use a less abundant isotopologue?,Drug Testing and Analysis - X-MOL

Expanding the linear dynamic range for quantitative liquid chromatography-high resolution mass spectrometry utilizing natural isotopologue signals - ScienceDirect

Figure 1 from Mass Spectrometry-Based Identification of Muscle-Associated and Muscle-Derived Proteomic Biomarkers of Dystrophinopathies. | Semantic Scholar

Protein proportions and dynamic range in murine skeletal muscle tissue.... | Download Scientific Diagram

3.3. Estimating the linear range | MOOC: Validation of liquid chromatography mass spectrometry (LC-MS) methods (analytical chemistry) course

Expanding the linear dynamic range for quantitative liquid chromatography-high resolution mass spectrometry utilizing natural isotopologue signals - ScienceDirect
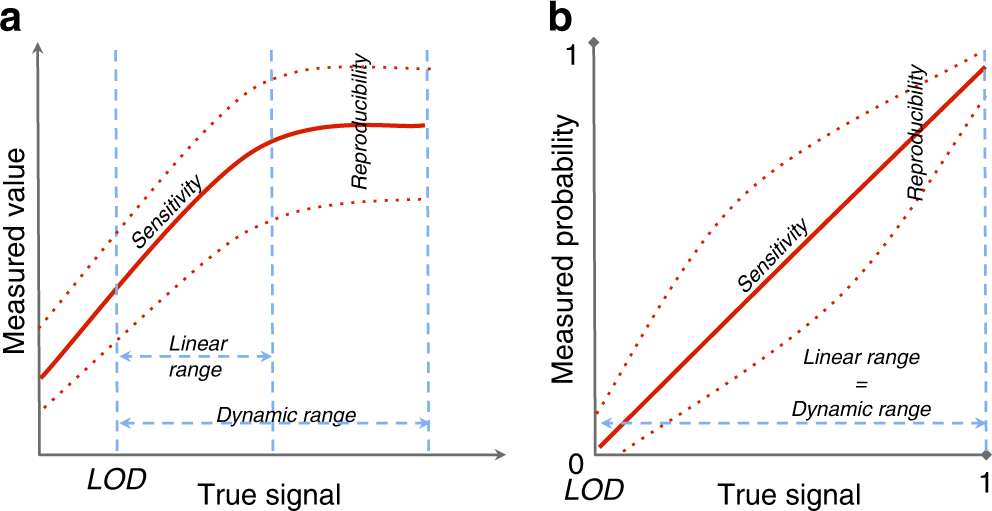
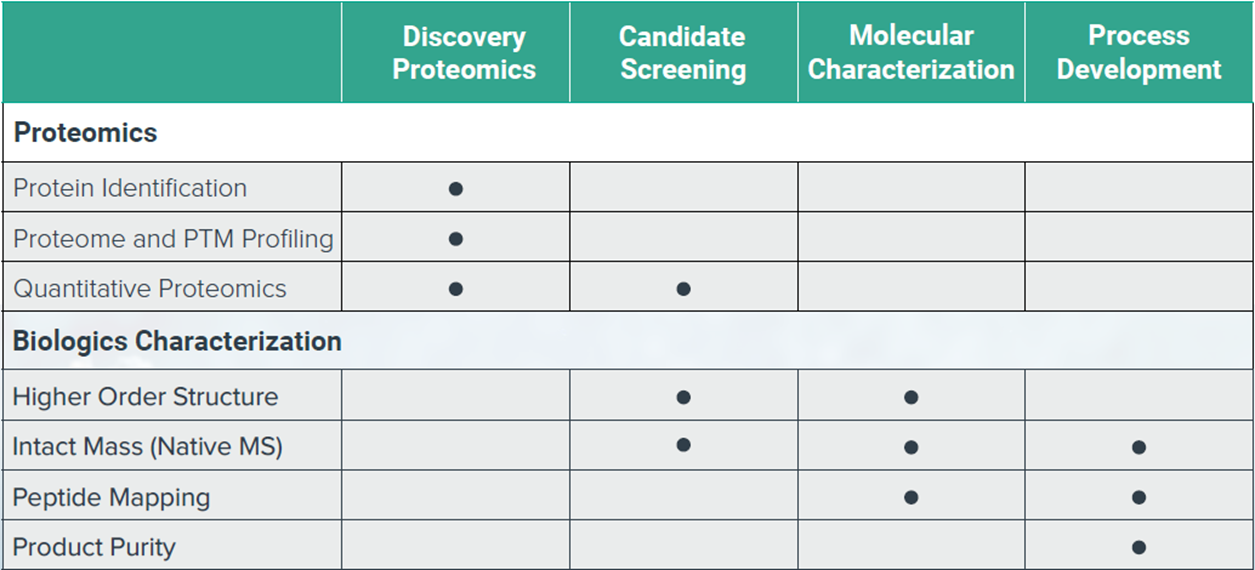
Harmonization of quality metrics and power calculation in multi-omic studies | Nature Communications

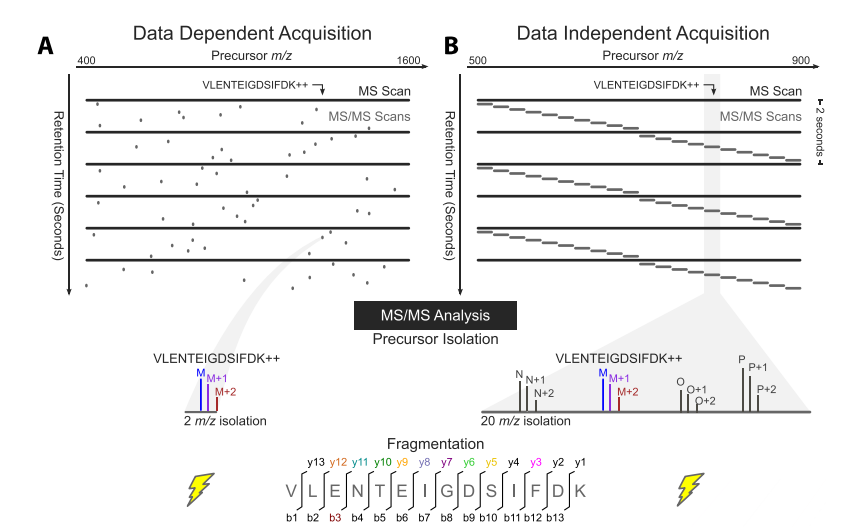
Assessing the Relationship Between Mass Window Width and Retention Time Scheduling on Protein Coverage for Data-Independent Acquisition | Journal of The American Society for Mass Spectrometry
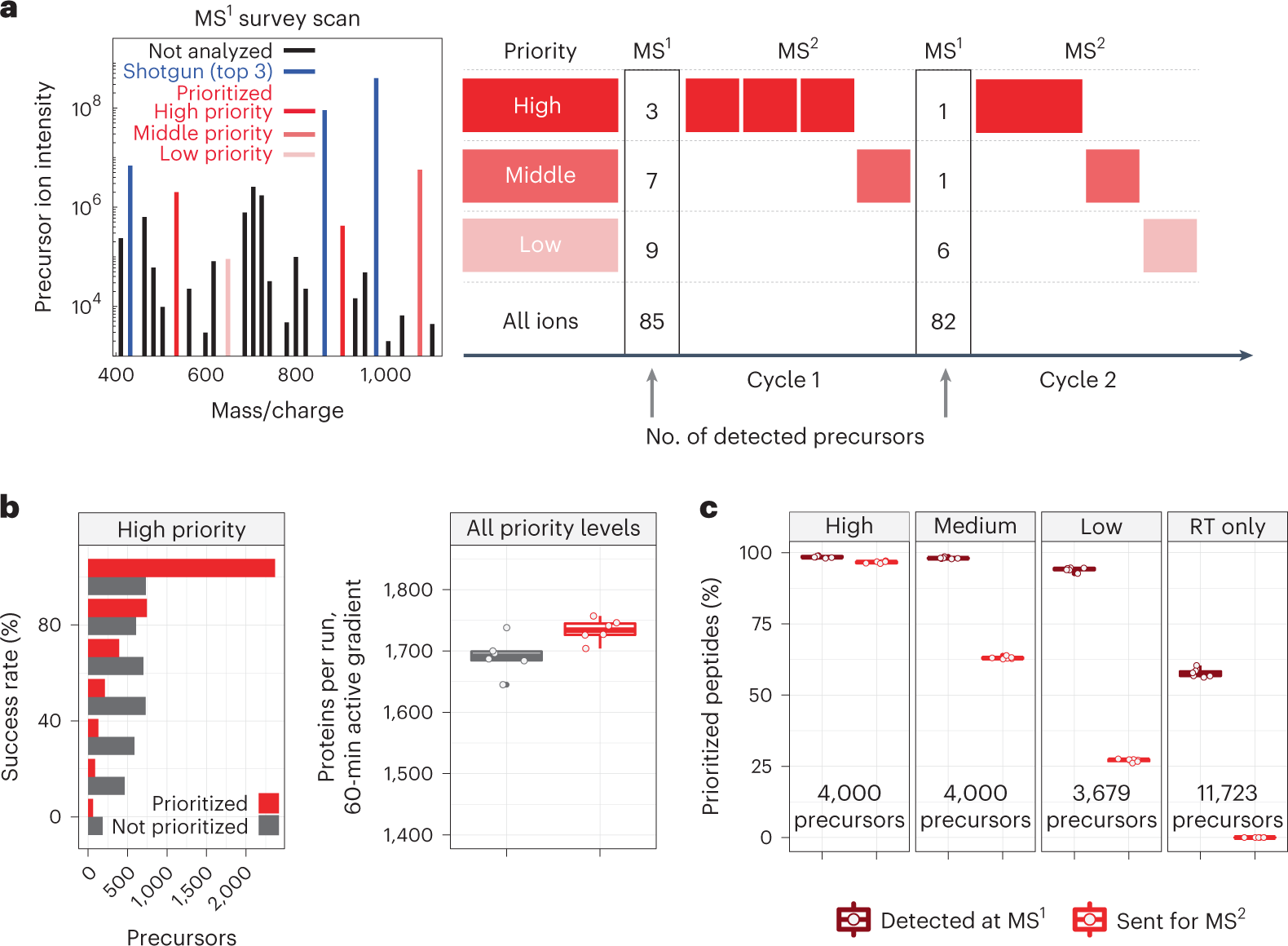
Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics | Nature Methods